Convert RGB three band to single band colour table
SGDF2PCT.RdThis function converts a three-band SpatialGridDataFrame into a single band of colour indices and a colour look-up table using RGB2PCT. vec2RGB uses given breaks and colours (like image) to make a three column matrix of red, green, and blue values for a numeric vector.
SGDF2PCT(x, ncolors = 256, adjust.bands=TRUE)
vec2RGB(vec, breaks, col)Arguments
- x
a three-band SpatialGridDataFrame object
- ncolors
a number of colours between 2 and 256
- adjust.bands
default TRUE; if FALSE the three bands must lie each between 0 and 255, but will not be streched within those bounds
- vec
a numeric vector
- breaks
a set of breakpoints for the colours: must give one more breakpoint than colour
- col
a list of colors
Value
The value returned is a list:
- idx
a vector of colour indices in the same spatial order as the input object
- ct
a vector of RGB colours
References
Examples
logo <- system.file("pictures/Rlogo.jpg", package="rgdal")[1]
SGlogo <- readGDAL(logo)
#> Warning: GDAL support is provided by the sf and terra packages among others
#> /tmp/RtmpnkEu0w/temp_libpath101e75200293a/rgdal/pictures/Rlogo.jpg has GDAL driver JPEG
#> and has 175 rows and 200 columns
#> Warning: GDAL support is provided by the sf and terra packages among others
#> Warning: GeoTransform values not available
cols <- SGDF2PCT(SGlogo)
SGlogo$idx <- cols$idx
image(SGlogo, "idx", col=cols$ct)
 SGlogo <- readGDAL(logo)
#> Warning: GDAL support is provided by the sf and terra packages among others
#> /tmp/RtmpnkEu0w/temp_libpath101e75200293a/rgdal/pictures/Rlogo.jpg has GDAL driver JPEG
#> and has 175 rows and 200 columns
#> Warning: GDAL support is provided by the sf and terra packages among others
#> Warning: GeoTransform values not available
cols <- SGDF2PCT(SGlogo, ncolors=64)
SGlogo$idx <- cols$idx
image(SGlogo, "idx", col=cols$ct)
SGlogo <- readGDAL(logo)
#> Warning: GDAL support is provided by the sf and terra packages among others
#> /tmp/RtmpnkEu0w/temp_libpath101e75200293a/rgdal/pictures/Rlogo.jpg has GDAL driver JPEG
#> and has 175 rows and 200 columns
#> Warning: GDAL support is provided by the sf and terra packages among others
#> Warning: GeoTransform values not available
cols <- SGDF2PCT(SGlogo, ncolors=64)
SGlogo$idx <- cols$idx
image(SGlogo, "idx", col=cols$ct)
 SGlogo <- readGDAL(logo)
#> Warning: GDAL support is provided by the sf and terra packages among others
#> /tmp/RtmpnkEu0w/temp_libpath101e75200293a/rgdal/pictures/Rlogo.jpg has GDAL driver JPEG
#> and has 175 rows and 200 columns
#> Warning: GDAL support is provided by the sf and terra packages among others
#> Warning: GeoTransform values not available
cols <- SGDF2PCT(SGlogo, ncolors=8)
SGlogo$idx <- cols$idx
image(SGlogo, "idx", col=cols$ct)
SGlogo <- readGDAL(logo)
#> Warning: GDAL support is provided by the sf and terra packages among others
#> /tmp/RtmpnkEu0w/temp_libpath101e75200293a/rgdal/pictures/Rlogo.jpg has GDAL driver JPEG
#> and has 175 rows and 200 columns
#> Warning: GDAL support is provided by the sf and terra packages among others
#> Warning: GeoTransform values not available
cols <- SGDF2PCT(SGlogo, ncolors=8)
SGlogo$idx <- cols$idx
image(SGlogo, "idx", col=cols$ct)
 data(meuse.grid)
coordinates(meuse.grid) <- c("x", "y")
gridded(meuse.grid) <- TRUE
fullgrid(meuse.grid) <- TRUE
summary(meuse.grid$dist)
#> Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
#> 0.000 0.119 0.272 0.297 0.440 0.993 5009
opar <- par(no.readonly=TRUE)
par(mfrow=c(1,2), mar=c(1,1,1,1)+0.1)
image(meuse.grid, "dist", breaks=seq(0,1,1/10), col=bpy.colors(10))
RGB <- vec2RGB(meuse.grid$dist, breaks=seq(0,1,1/10), col=bpy.colors(10))
summary(RGB)
#> red green blue
#> Min. : 0.00 Min. : 0.00 Min. : 36.0
#> 1st Qu.: 0.00 1st Qu.: 0.00 1st Qu.: 87.0
#> Median : 0.00 Median : 0.00 Median :189.0
#> Mean : 75.31 Mean : 22.98 Mean :171.1
#> 3rd Qu.:159.00 3rd Qu.: 15.00 3rd Qu.:255.0
#> Max. :255.00 Max. :255.00 Max. :255.0
#> NA's :5009 NA's :5009 NA's :5009
meuse.grid$red <- RGB[,1]
meuse.grid$green <- RGB[,2]
meuse.grid$blue <- RGB[,3]
cols <- SGDF2PCT(meuse.grid[c("red", "green", "blue")], ncolors=10,
adjust.bands=FALSE)
is.na(cols$idx) <- is.na(meuse.grid$dist)
meuse.grid$idx <- cols$idx
image(meuse.grid, "idx", col=cols$ct)
data(meuse.grid)
coordinates(meuse.grid) <- c("x", "y")
gridded(meuse.grid) <- TRUE
fullgrid(meuse.grid) <- TRUE
summary(meuse.grid$dist)
#> Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
#> 0.000 0.119 0.272 0.297 0.440 0.993 5009
opar <- par(no.readonly=TRUE)
par(mfrow=c(1,2), mar=c(1,1,1,1)+0.1)
image(meuse.grid, "dist", breaks=seq(0,1,1/10), col=bpy.colors(10))
RGB <- vec2RGB(meuse.grid$dist, breaks=seq(0,1,1/10), col=bpy.colors(10))
summary(RGB)
#> red green blue
#> Min. : 0.00 Min. : 0.00 Min. : 36.0
#> 1st Qu.: 0.00 1st Qu.: 0.00 1st Qu.: 87.0
#> Median : 0.00 Median : 0.00 Median :189.0
#> Mean : 75.31 Mean : 22.98 Mean :171.1
#> 3rd Qu.:159.00 3rd Qu.: 15.00 3rd Qu.:255.0
#> Max. :255.00 Max. :255.00 Max. :255.0
#> NA's :5009 NA's :5009 NA's :5009
meuse.grid$red <- RGB[,1]
meuse.grid$green <- RGB[,2]
meuse.grid$blue <- RGB[,3]
cols <- SGDF2PCT(meuse.grid[c("red", "green", "blue")], ncolors=10,
adjust.bands=FALSE)
is.na(cols$idx) <- is.na(meuse.grid$dist)
meuse.grid$idx <- cols$idx
image(meuse.grid, "idx", col=cols$ct)
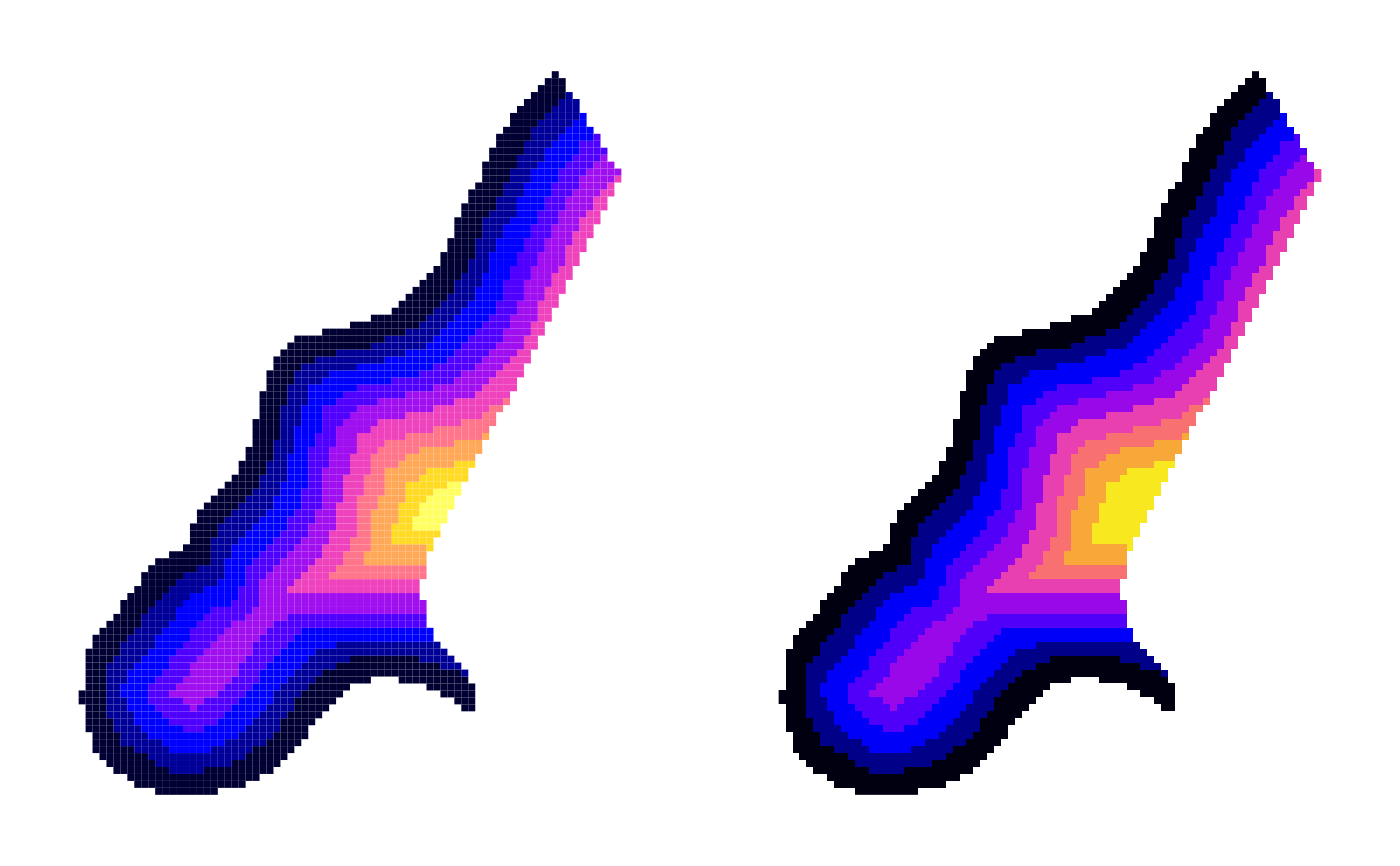 par(opar)
# Note: only one wrongly classified pixel after NA handling/dropping
# The functions are not written to be reversible
sort(table(findInterval(meuse.grid$dist, seq(0,1,1/10), all.inside=TRUE)))
#>
#> 10 9 8 7 6 5 4 2 3 1
#> 28 54 99 118 237 417 431 507 525 687
sort(table(cols$idx))
#>
#> 10 1 2 3 4 6 5 9 7
#> 82 99 118 237 417 431 507 525 687
par(opar)
# Note: only one wrongly classified pixel after NA handling/dropping
# The functions are not written to be reversible
sort(table(findInterval(meuse.grid$dist, seq(0,1,1/10), all.inside=TRUE)))
#>
#> 10 9 8 7 6 5 4 2 3 1
#> 28 54 99 118 237 417 431 507 525 687
sort(table(cols$idx))
#>
#> 10 1 2 3 4 6 5 9 7
#> 82 99 118 237 417 431 507 525 687